
- #Phyton data universal database how to#
- #Phyton data universal database install#
- #Phyton data universal database download#
Vincent Scalfani, Chemistry and Informatics Librarian, Rodgers Library for Science and Engineering. Keep API Keys private to you only (i.e., treat it as a password). Please try and pre-register for a Scopus API Key. An introductory workshop will be offered on Thursday, September 22 at 3:00 pm in the Bruno Library. Some prior experience with Python is recommended. Use the Pandas library to compile and perform basic data analysis. Utilize loops to automate data extraction. In addition to working with the Pybliometrics wrapper for accessing the Scopus API, participants will learn to do the following: Participants in this workshop utilize Python to retrieve citation data from the Scopus API. Mode: In-person, Rodgers Library, Scholar’s Stationĭate: Thursday, September 29, 3:00-4:45 PM You may need to read files or csv files, these articles will help you: Read file Write. If you are new to databases, I recommend one of these courses: Master SQL Databases with Python Python and (Relational) Database Systems: SQLite MySQL PostgreSQL ORM with SqlAlchemy Raw data. Using Python to Search and Retrieve Data from the Elsevier Scopus Database Exploring a Sqlite database with sqliteman. Location: Scholars' Station - Rodgers Library Presenter: Vincent Scalfani & Lance Simpson
#Phyton data universal database how to#
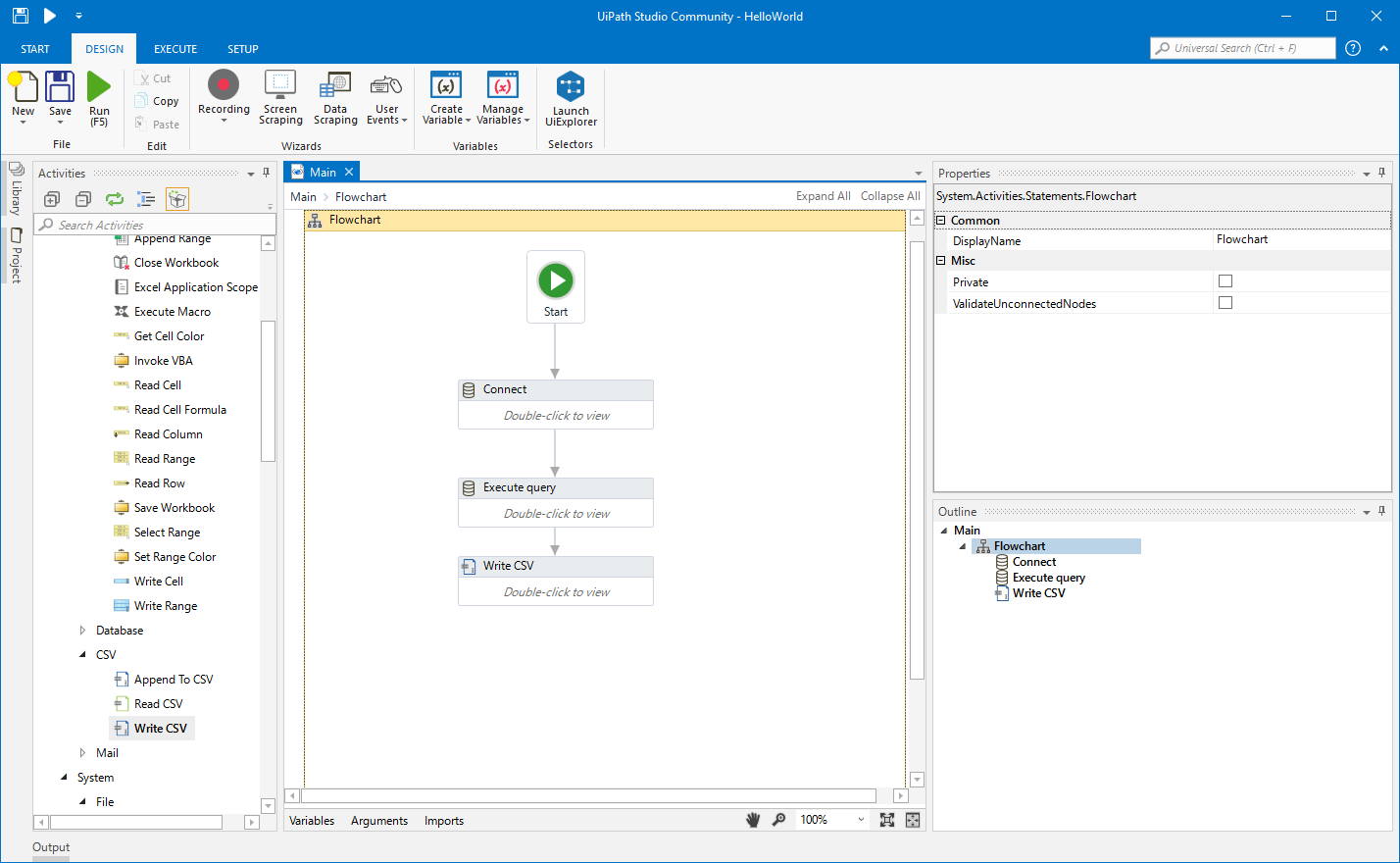
The following code blocks demonstrate how to get datetimes and numeric values from the retrieved string representations (note the use of the use of the pipes). # Query for patients allergic to Pulse Vegetable query_string = ' PREFIX rdf: PREFIX rdfs: PREFIX sphn: PREFIX resource: PREFIX xsd: PREFIX snomed: SELECT DISTINCT ?patient WHERE ' # run query and retrieve results query_results <- SPARQL ( endpoint, query_string, ns = prefixes ) json extension.) Note that dump () takes two positional arguments: (1) the data object to be serialized, and (2) the file-like object to which the bytes will be written. Pip package in the current Jupyter kernel and import the module in Python: Using Python’s context manager, you can create a file called datafile.json and open it in write mode.
#Phyton data universal database install#
The following code example shows how to install a The SPARQL Endpoint interface to Python ‘SPARQLWrapper’ In order to access the RDF data loaded in GraphDB from Python, Loading data from GraphDB in Python and R Step 1: Handle dependencies Python Table 1: Overview of the employed languages, development environments, and graph database. For installation guides please refer to the documentation provided in the following links: This section tackles different languages and development environments which are summarized in Table 1. Two programmating languages: R and Python. It showcases how RDF graphs can be queries and ‘manipulated’ with Interested in analysis their data through other means than using a triplestore. This document is mainly intended for researchers and RDF experts who are The instructions presented in this page have been integrated in a notebook. (learn more about Query data with SPARQL). The SPARQL queries employed in these examples build upon

Introduced in the “Graphical exploration of data with GraphDB” section The examples used in this page are based on the mock-data How to use Python and R with RDF data Training External Terminologies in RDF for projects.
#Phyton data universal database download#
Download external terminologies from the Terminology Service.Dealing with various datatypes (datetime, numeric, etc.).


 0 kommentar(er)
0 kommentar(er)
